- The full article entitled “Energy Counterbalance to Harness Photoinduced Structural Planarization of Dibenzo[b,f]azepines toward Thermal Reversibility” can be found at the JACS website at https://pubs.acs.org/doi/10.1021/jacs.1c11231
- Authors: Yi Chen, Sheng-Ming Tseng, Kai-Hsin Chang, Pi-Tai Chou*
In sharp contrast to most photoinduced structural planarization (PISP) phenomena, which are highly exergonic and irreversible processes (see PDBA in Figure 1a as an example), we report here a series of a new class of PISP molecules, 9-phenyl-9H-tribenzo[b,d,f]azepine (PTBA) and its derivatives, where PISP is within the thermally reversible regime (see Figure 1b). The underlying foundation is the energy counterbalance along PISP, where upon electronic excitation the azepine core chromophore undergoes planarization to gain stabilization by a cyclic 4n π conjugation (n: integer number, according to the Baird’s rule). Concurrently, the C7=C8 fused benzene ring of PTBA is prone to gain aromaticity, which conversely decreases the 4n π-electron resonance stabilization of the 9H-tribenzo[b,d,f]azepine. The offset results in a minimum energy state (P*) along PISP that is in thermal equilibrium with the initially prepared state (R*). The relaxed structure of R* deviates greatly from the planar configuration commonly seen in PISP. PISP of PTBAs is thus sensitive to the solvent polarity, temperature and substituents, causing prominent stimuli-dependent ratiometric fluorescence for R* versus P*. The exploitation of the energy counterbalance effect proves to be a practical strategy for harnessing excited-state structural relaxation.
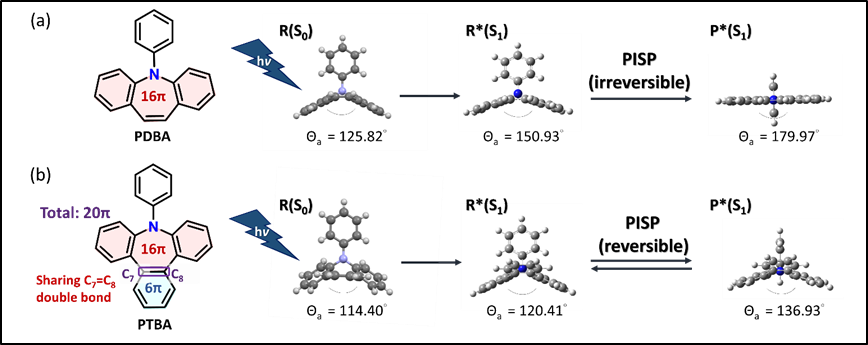
Figure 1. Illustration of irreversible and reversible PISP using (a)PDBA and (b)PTBA, respectively, as examples. PTBA is the core structure in this study, while the excited-state structures of PDBA and PTBA in R*(S1) and P*(S1) are calculated at (TD)PBE0/6-31+G(d,p) level. Both structures of PDBA and PTBA in R(S0) are extracted from single crystal X-ray diffraction data.